| Publication |
|
Tags |
| 2024 |

|
Junxiong Wang, Tushaar Gangavarapu, Jing Nathan Yan, and Alexander M. Rush (2024): MambaByte: Token-free Selective State Space Model. In Proceedings of the Conference on Language Modeling (COLM 2024).
abstract
x
MambaByte: Token-free Selective State Space Model
Token-free language models learn directly from raw bytes and remove the inductive bias of subword tokenization. Operating on bytes, however, results in significantly longer sequences. In this setting, standard autoregressive Transformers scale poorly as the effective memory required grows with sequence length. The recent development of the Mamba state space model (SSM) offers an appealing alternative approach with a fixed-sized memory state and efficient decoding. We propose MambaByte, a token-free adaptation of the Mamba SSM trained autoregressively on byte sequences. In terms of modeling, we show MambaByte to be competitive with, and even to outperform, state-of-the-art subword Transformers on language modeling tasks while maintaining the benefits of token-free language models, such as robustness to noise. In terms of efficiency, we develop an adaptation of speculative decoding with tokenized drafting and byte-level verification. This results in a 2.6× inference speedup to the standard MambaByte implementation, showing similar decoding efficiency as the subword Mamba. These findings establish the viability of SSMs in enabling token-free language modeling.
url
.pdf
.bib
x
MambaByte: Token-free Selective State Space Model
@misc{gangavarapu-mambabyte-2024,
title="MambaByte: Token-free Selective State Space Model",
author="Wang, Junxiong and Gangavarapu, Tushaar and Jing, Nathan Yan and Rush, Alexander M.",
journal="arXiv preprint arXiv:2401.13660",
year="2024",
publisher="arXiv",
url=""
}
models
video
|
conference
state space models
mamba
llm
language modeling
byte-level models
|
| 2023 |
|
Yann Hicke, Abhishek Masand, Wentao Guo, and Tushaar Gangavarapu (2023): Assessing the efficacy of large language models in generating accurate teacher responses. In Proceedings of the 18th Workshop on Innovative Use of NLP for Building Educational Applications (BEA 2023).
abstract
x
Assessing the efficacy of large language models in generating accurate teacher responses
(Tack et al., 2023) organized the shared task hosted by the 18th Workshop on Innovative Use of NLP for Building Educational Applications on generation of teacher language in educational dialogues. Following the structure of the shared task, in this study, we attempt to assess the generative abilities of large language models in providing informative and helpful insights to students, thereby simulating the role of a knowledgeable teacher. To this end, we present an extensive evaluation of several benchmarking generative models, including GPT-4 (few-shot, in-context learning), fine-tuned GPT-2, and fine-tuned DialoGPT. Additionally, to optimize for pedagogical quality, we fine-tuned the Flan-T5 model using reinforcement learning. Our experimental findings on the Teacher-Student Chatroom Corpus subset indicate the efficacy of GPT-4 over other fine-tuned models, measured using BERTScore and DialogRPT. We hypothesize that several dataset characteristics, including sampling, representativeness, and dialog completeness, pose significant challenges to fine-tuning, thus contributing to the poor generalizability of the fine-tuned models. Finally, we note the need for these generative models to be evaluated with a metric that relies not only on dialog coherence and matched language modeling distribution but also on the model’s ability to showcase pedagogical skills.
url
.pdf
.bib
x
Assessing the efficacy of large language models in generating accurate teacher responses
@inproceedings{gangavarapu-bea-2023,
title="Assessing the efficacy of large language models in generating accurate teacher responses",
author="Hicke, Yann and Masand, Abhishek and Guo, Wentao and Gangavarapu, Tushaar",
booktitle="Proceedings of the 18th Workshop on Innovative Use of NLP for Building Educational Applications (BEA 2023)",
year="2023",
publisher="Association for Computational Linguistics",
url="https://aclanthology.org/2023.bea-1.60"
}
code
shared task
|
conference
ai for education
llms
finetuning
efficacy
student-teacher interaction
llm prompting
|
| 2022 |

|
Tushaar Gangavarapu and Sriraghavendra Ramaswamy (2022): Alexa, stop reading the references: Enhancing the reading experience in Kindle eBooks. In Proceedings of the 9th Amazon Machine Learning Conference (AMLC), Amazon.
abstract
x
Alexa, stop reading the references: Enhancing the reading experience in Kindle eBooks.
NOTE: Owing to the confidentiality policy at Amazon, the abstract of this paper is not published here. But, if you are an Amazon employee, please visit the AMLC 2022 conference site (or alternatively, use the "url" or ".pdf" links indicated on the webpage) for the specifics of this research. (If, however, you are sriragha@ or anudee@, then I'm honored that you, the legend of legends, visited my page!) Anyway, please feel free to reach out to sriragha@amazon.com (or medhini@amazon.com) for details on the conducted research.
url
.pdf
.bib
x
Alexa, stop reading the references: Enhancing the reading experience in Kindle eBooks.
@inproceedings{gangavarapu-amlc-2022a,
title="Alexa, stop reading the references: Enhancing the reading experience in Kindle eBooks",
author="Gangavarapu, Tushaar and Ramaswamy, Sriraghavendra",
booktitle="Proceedings of the 9th Amazon Machine Learning Conference (AMLC)",
year="2022",
publisher="Amazon"
}
|
conference
alexa
ebooks
kindle
natural language processing
reading experience
user engagement
|
|
Tushaar Gangavarapu* and Sriraghavendra Ramaswamy* (2022): A figure is worth a thousand words, but where are the words?: Enhancing image experience in Kindle eBooks. In Proceedings of the 9th Amazon Machine Learning Conference (AMLC), Amazon.
abstract
x
A figure is worth a thousand words, but where are the words?: Enhancing image experience in Kindle eBooks.
NOTE: Owing to the confidentiality policy at Amazon, the abstract of this paper is not published here. But, if you are an Amazon employee, please visit the AMLC 2022 conference site (or alternatively, use the "url" or ".pdf" links indicated on the webpage) for the specifics of this research. (If, however, you are sriragha@ or anudee@, then I'm honored that you, the legend of legends, visited my page!) Anyway, please feel free to reach out to sriragha@amazon.com (or medhini@amazon.com) for details on the conducted research.
url
.pdf
.bib
x
A figure is worth a thousand words, but where are the words?: Enhancing image experience in Kindle eBooks.
@inproceedings{gangavarapu-amlc-2022b,
title="A figure is worth a thousand words, but where are the words?: Enhancing image experience in Kindle eBooks",
author="Gangavarapu, Tushaar and Ramaswamy, Sriraghavendra",
booktitle="Proceedings of the 9th Amazon Machine Learning Conference (AMLC)",
year="2022",
publisher="Amazon"
}
|
conference
ebooks
image processing
kindle
natural language processing
reading experience
user engagement
|
| 2021 |

|
Tushaar Gangavarapu, Gokul S Krishnan, Sowmya Kamath S, and Jayakumar Jeganathan (2021): FarSight: Long-Term Disease Prediction Using Unstructured Clinical Nursing Notes. Transactions on Emerging Topics in Computing (TETC), IEEE.
abstract
x
FarSight: Long-Term Disease Prediction Using Unstructured Clinical Nursing Notes.
Accurate risk stratification using patient data is a vital task in channeling prioritized care. Most state-of-the-art models are predominantly reliant on digitized data in the form of structured Electronic Health Records (EHRs). Those models overlook the valuable patient-specific information embedded in unstructured clinical notes, which is the prevalent medium employed by caregivers to record patients' disease timeline. The availability of such patient-specific data presents an unprecedented opportunity to build intelligent systems that provide exclusive insights into patients' disease physiology. Moreover, very few works have attempted to benchmark the performance of deep neural architectures against the state-of-the-art models on publicly available datasets. This paper presents significant observations from our benchmarking experiments on the applicability of deep learning models for the clinical task of ICD-9 code group prediction. We present FarSight, a long-term aggregation mechanism intended to recognize the onset of the disease with the earliest detected symptoms. Vector space and topic modeling approaches are utilized to capture the semantic information in the patient representations. Experiments on MIMIC-III database underscored the superior performance of the proposed models built on unstructured data when compared to structured EHR based state-of-the-art model, achieving an improvement of 19.34% in AUPRC and 5.41% in AUROC.
url
.pdf (draft)
.bib
x
FarSight: Long-Term Disease Prediction Using Unstructured Clinical Nursing Notes.
@article{gangavarapu-tetc-2021,
author="Gangavarapu, Tushaar and Krishnan, Gokul S and Kamath S, Sowmya and Jeganathan, Jayakumar",
title="FarSight: Long-Term Disease Prediction Using Unstructured Clinical Nursing Notes",
journal="IEEE Transactions on Emerging Topics in Computing",
doi="10.1109/TETC.2020.2975251",
volume="9",
number="3",
pages="1151-1169",
year="2021"
}
.data
|
journal
clinical decision support systems
disease prediction
healthcare analytics
ICD-9 code group prediction
precision medicine
|

|
Veena Mayya, Sowmya Kamath S, Gokul S Krishnan, and Tushaar Gangavarapu (2021): Multi-channel, convolutional attention based neural model for automated diagnostic coding of unstructured patient discharge summaries. Future Generation Computer Systems (FGCS), Elsevier.
abstract
x
Multi-channel, convolutional attention based neural model for automated diagnostic coding of unstructured patient discharge summaries.
Effective coding of patient records in hospitals is an essential requirement for epidemiology, billing, and managing insurance claims. The prevalent practice of manual coding, carried out by trained medical coders, is error-prone and time-consuming. Mitigating this labor-intensive process by developing diagnostic coding systems built on patients' Electronic Medical Records (EMRs) is vital. However, developing nations with low digitization rates have limited availability of structured EMRs, thereby demanding the need for systems built on unstructured data sources. Despite the rich clinical information available in such unstructured data, modeling them is complex, owing to the variety and sparseness of diagnostic codes, complex structural and temporal nature of summaries, and prolific use of medical jargon. This work proposes a context-attentive network to facilitate automatic diagnostic code assignment as a multi-label classification problem. The proposed model facilitates information aggregation across a patient's discharge summary via multi-channel, variable-sized convolutional filters to extract multi-granular snippets. The attention mechanism enables selecting vital segments in those snippets that map to the clinical codes. The model's superior performance underscores its effectiveness compared to the state-of-the-art on the MIMIC-III database. Additionally, experimental validation using the CodiEsp dataset exhibited the model's interpretability and explainability.
url
.pdf (draft)
.highlights
.bib
x
@article{gangavarapu-fgcs-2021,
author="Mayya, Veena and Kamath S, Sowmya and Krishnan, Gokul S and Gangavarapu, Tushaar",
title="Multi-channel, convolutional attention based neural model for automated diagnostic coding of unstructured patient discharge summaries",
journal="Future Generation Computer Systems",
year="2021",
issn="0167-739X",
doi="10.1016/j.future.2021.01.013",
url="http://www.sciencedirect.com/science/article/pii/S0167739X21000236"
}
.data
|
journal
disease prediction
explainability
healthcare informatics
interpretability
predictive analytics
unstructured text modeling
|
| 2020 |
|
Tushaar Gangavarapu and Jaidhar CD (2020): A Novel Bio-inspired Hybrid Metaheuristic for Unsolicited Bulk Email Detection. In Proceedings of the 20th International Conference on Computational Science (ICCS), Springer (LNCS).
abstract
x
A Novel Bio-inspired Hybrid Metaheuristic for Unsolicited Bulk Email Detection.
With the recent influx of technology, Unsolicited Bulk Emails (UBEs) have become a potential problem, leaving computer users and organizations at the risk of brand, data, and financial loss. In this paper, we present a novel bio-inspired hybrid parallel optimization algorithm (Cuckoo-Firefly-GR), which combines Genetic Replacement (GR) of low fitness individuals with a hybrid of Cuckoo Search (CS) and Firefly (FA) optimizations. Cuckoo-Firefly-GR not only employs the random walk in CS, but also uses mechanisms in FA to generate and select fitter individuals. The content- and behavior-based features of emails used in the existing works, along with Doc2Vec features of the email body are employed to extract the syntactic and semantic information in the emails. By establishing an optimal balance between intensification and diversification, and reaching global optimization using two metaheuristics, we argue that the proposed algorithm significantly improves the performance of UBE detection, by selecting the most discriminative feature subspace. This study presents significant observations from the extensive evaluations on UBE corpora of 3,844 emails, that underline the efficiency and superiority of our proposed Cuckoo-Firefly-GR over the base optimizations (Cuckoo-GR and Firefly-GR), dense autoencoders, recurrent neural autoencoders, and several state-of-the-art methods. Furthermore, the instructive feature subset obtained using the proposed Cuckoo-Firefly-GR, when classified using a dense neural model, achieved an accuracy of 99%.
url
.pdf (draft)
.bib
x
A Novel Bio-inspired Hybrid Metaheuristic for Unsolicited Bulk Email Detection.
@inproceedings{gangavarapu-iccs-2020,
title="A Novel Bio-inspired Hybrid Metaheuristic for Unsolicited Bulk Email Detection",
author="Gangavarapu, Tushaar and Jaidhar, CD",
booktitle="Computational Science – ICCS 2020",
year="2020",
publisher="Springer International Publishing",
address="Cham"
}
.data
|
conference
evolutionary computing
feature selection
internet security
metaheuristics
natural language processing
phishing
spam
|

|
Tushaar Gangavarapu, Jaidhar CD, and Bhabesh Chanduka (2020): Applicability of machine learning in spam and phishing email filtering: review and approaches. Artificial Intelligence Review (AIRE), Springer Nature.
abstract
x
Applicability of machine learning in spam and phishing email filtering: review and approaches.
With the influx of technological advancements and the increased simplicity in communication, especially through emails, the upsurge in the volume of Unsolicited Bulk Emails (UBEs) has become a severe threat to global security and economy. Spam emails not only waste users' time, but also consume a lot of network bandwidth, and may also include malware as executable files. Alternatively, phishing emails falsely claim users' personal information to facilitate identity theft and are comparatively more dangerous. Thus, there is an intrinsic need for the development of more robust and dependable UBE filters that facilitate automatic detection of such emails. There are several countermeasures to spam and phishing, including blacklisting and content-based filtering. However, in addition to content-based features, behavior-based features are well-suited in the detection of UBEs. Machine learning models are being extensively used by leading internet service providers like Yahoo, Gmail, and Outlook, to filter and classify UBEs successfully. There are far too many options to consider, owing to the need to facilitate UBE detection and the recent advances in this domain. In this paper, we aim at elucidating on the way of extracting email content and behavior-based features, what features are appropriate in the detection of UBEs, and the selection of the most discriminating feature set. Furthermore, to accurately handle the menace of UBEs, we facilitate an exhaustive comparative study using several state-of-the-art machine learning algorithms. Our proposed models resulted in an overall accuracy of 99% in the classification of UBEs. The text is accompanied by snippets of Python code, to enable the reader to implement the approaches elucidated in this paper.
url
.pdf (draft)
.bib
x
Applicability of machine learning in spam and phishing email filtering: review and approaches.
@article{gangavarapu-aire-2020,
author="Gangavarapu, Tushaar and Jaidhar, CD and Chanduka, Bhabesh",
title="Applicability of machine learning in spam and phishing email filtering: review and approaches",
journal="Artificial Intelligence Review",
issn="0269–2821",
doi="10.1007/s10462-020-09814-9",
year="2020"
}
code
.data
|
journal
feature engineering
machine learning
phishing
Python
spam
|
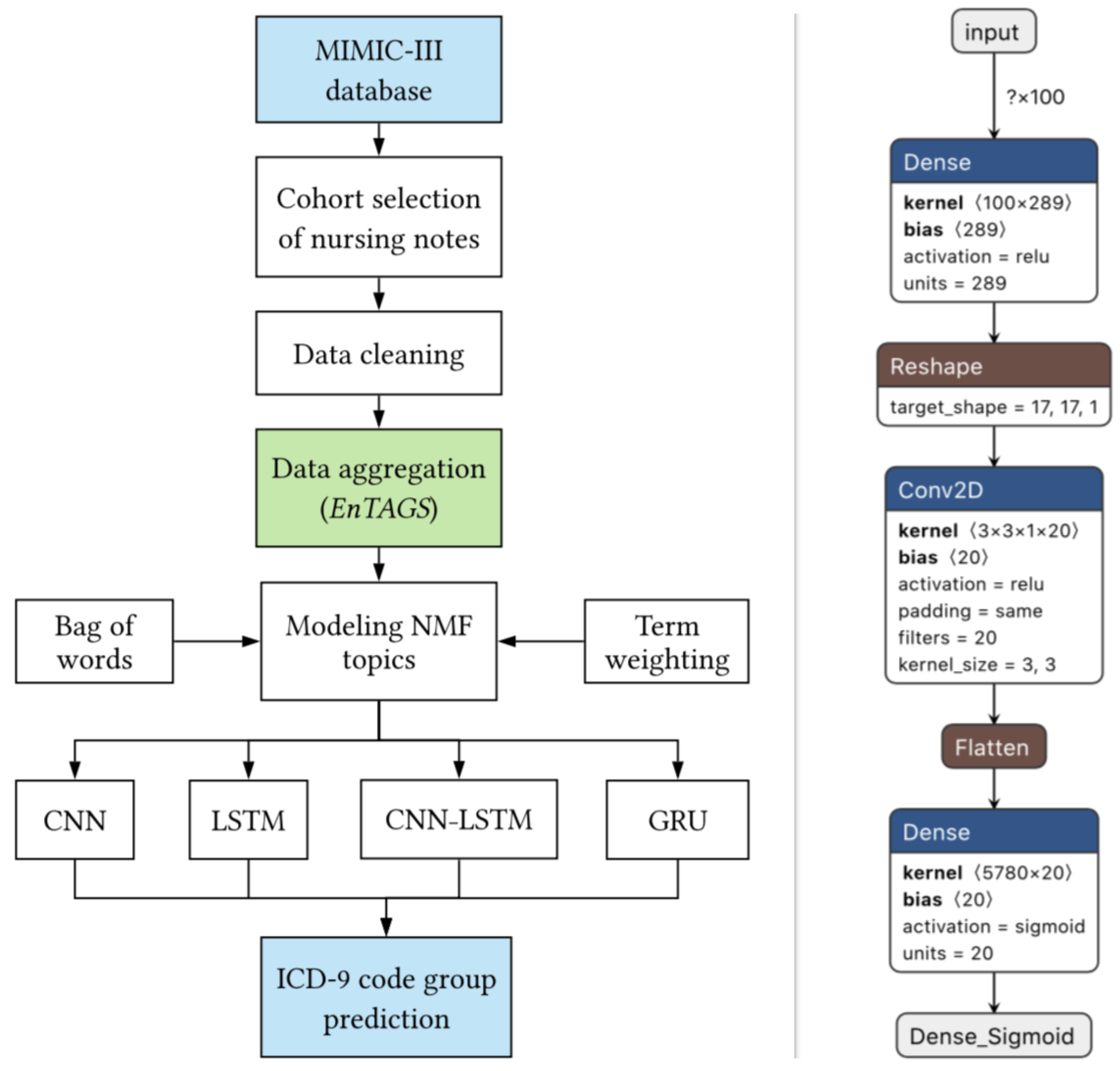
|
Aditya Jayasimha*, Tushaar Gangavarapu*, Sowmya Kamath S, and Gokul S Krishnan (2020): Deep Neural Learning for Automated Diagnostic Code Group Prediction Using Unstructured Nursing Notes. In Proceedings of the ACM India Joint International Conference on Data Science and Management of Data (7th ACM IKDD CoDS and 25th COMAD), ACM (pp. 152–160).
abstract
x
Deep Neural Learning for Automated Diagnostic Code Group Prediction Using Unstructured Nursing Notes.
Disease prediction, a central problem in clinical care and management, has gained much significance over the last decade. Nursing notes documented by caregivers contain valuable information concerning a patient's state, which can aid in the development of intelligent clinical prediction systems. Moreover, due to the limited adaptation of structured electronic health records in developing countries, the need for disease prediction from such clinical text has garnered substantial interest from the research community. The availability of large, publicly available databases such as MIMIC-III, and advancements in machine and deep learning models with high predictive capabilities have further facilitated research in this direction. In this work, we model the latent knowledge embedded in the unstructured clinical nursing notes, to address the clinical task of disease prediction as a multi-label classification of ICD-9 code groups. We present EnTAGS, which facilitates aggregation of the data in the clinical nursing notes of a patient, by modeling them independent of one another. To handle the sparsity and high dimensionality of clinical nursing notes effectively, our proposed EnTAGS is built on the topics extracted using Non-negative matrix factorization. Furthermore, we explore the applicability of deep learning models for the clinical task of disease prediction, and assess the reliability of the proposed models using standard evaluation metrics. Our experimental evaluation revealed that the proposed approach consistently exceeded the state-of-the-art prediction model by 1.87% in accuracy, 12.68% in AUPRC, and 11.64% in MCC score.
url
.pdf
.poster
.bib
x
Deep Neural Learning for Automated Diagnostic Code Group Prediction Using Unstructured Nursing Notes.
@inproceedings{gangavarapu-cods-comad-2020,
title="Deep Neural Learning for Automated Diagnostic Code Group Prediction Using Unstructured Nursing Notes",
author="Jayasimha, Aditya and Gangavarapu, Tushaar and Kamath S, Sowmya and Krishnan, Gokul S",
booktitle="Proceedings of the ACM India Joint International Conference on Data Science and Management of Data",
series = "CoDS-COMAD '20",
year="2020",
publisher="ACM",
address = "New York, NY, USA",
pages="152-160",
numpages = "9",
doi="10.1145/3371158.3371176",
location="Hyderabad, India"
}
.data
|
conference
clinical decision support systems
deep learning
disease prediction
healthcare analytics
multi-label classification
natural language processing
|
| 2019 |

|
Tushaar Gangavarapu, Aditya Jayasimha, Gokul S Krishnan, and Sowmya Kamath S (2019): Predicting ICD-9 code groups with fuzzy similarity based supervised multi-label classification of unstructured clinical nursing notes. Knowledge-Based Systems (KnoSys), Elsevier 190:105321.
abstract
x
Predicting ICD-9 code groups with fuzzy similarity based supervised multi-label classification of unstructured clinical nursing notes.
In hospitals, caregivers are trained to chronicle the subtle changes in the clinical conditions of a patient at regular intervals, for enabling decision-making. Caregivers’ text-based clinical notes are a significant source of rich patient-specific data, that can facilitate effective clinical decision support, despite which, this treasure-trove of data remains largely unexplored for supporting the prediction of clinical outcomes. The application of sophisticated data modeling and prediction algorithms with greater computational capacity have made disease prediction from raw clinical notes a relevant problem. In this paper, we propose an approach based on vector space and topic modeling, to structure the raw clinical data by capturing the semantic information in the nursing notes. Fuzzy similarity based data cleansing approach was used to merge anomalous and redundant patient data. Furthermore, we utilize eight supervised multi-label classification models to facilitate disease (ICD-9 code group) prediction. We present an exhaustive comparative study to evaluate the performance of the proposed approaches using standard evaluation metrics. Experimental validation on MIMIC-III, an open database, underscored the superior performance of the proposed Term weighting of unstructured notes AGgregated using fuzzy Similarity (TAGS) model, which consistently outperformed the state-of-the-art structured data based approach by 7.79% in AUPRC and 1.24% in AUROC.
url
.pdf (draft)
.highlights
.bib
x
Predicting ICD-9 code groups with fuzzy similarity based supervised multi-label classification of unstructured clinical nursing notes.
@article{gangavarapu-knosys-2019,
author="Gangavarapu, Tushaar and Jayasimha, Aditya and Krishnan, Gokul S and Kamath S, Sowmya",
title="Predicting ICD-9 code groups with fuzzy similarity based supervised multi-label classification of unstructured clinical nursing notes",
journal="Knowledge-Based Systems",
pages="105321",
volume="190",
year="2019",
issn="0950-7051",
doi="10.1016/j.knosys.2019.105321",
url="http://www.sciencedirect.com/science/article/pii/S0950705119305982"
}
.data
|
journal
clinical decision support systems
disease prediction
healthcare analytics
ICD-9 code group prediction
machine learning
natural language processing
|
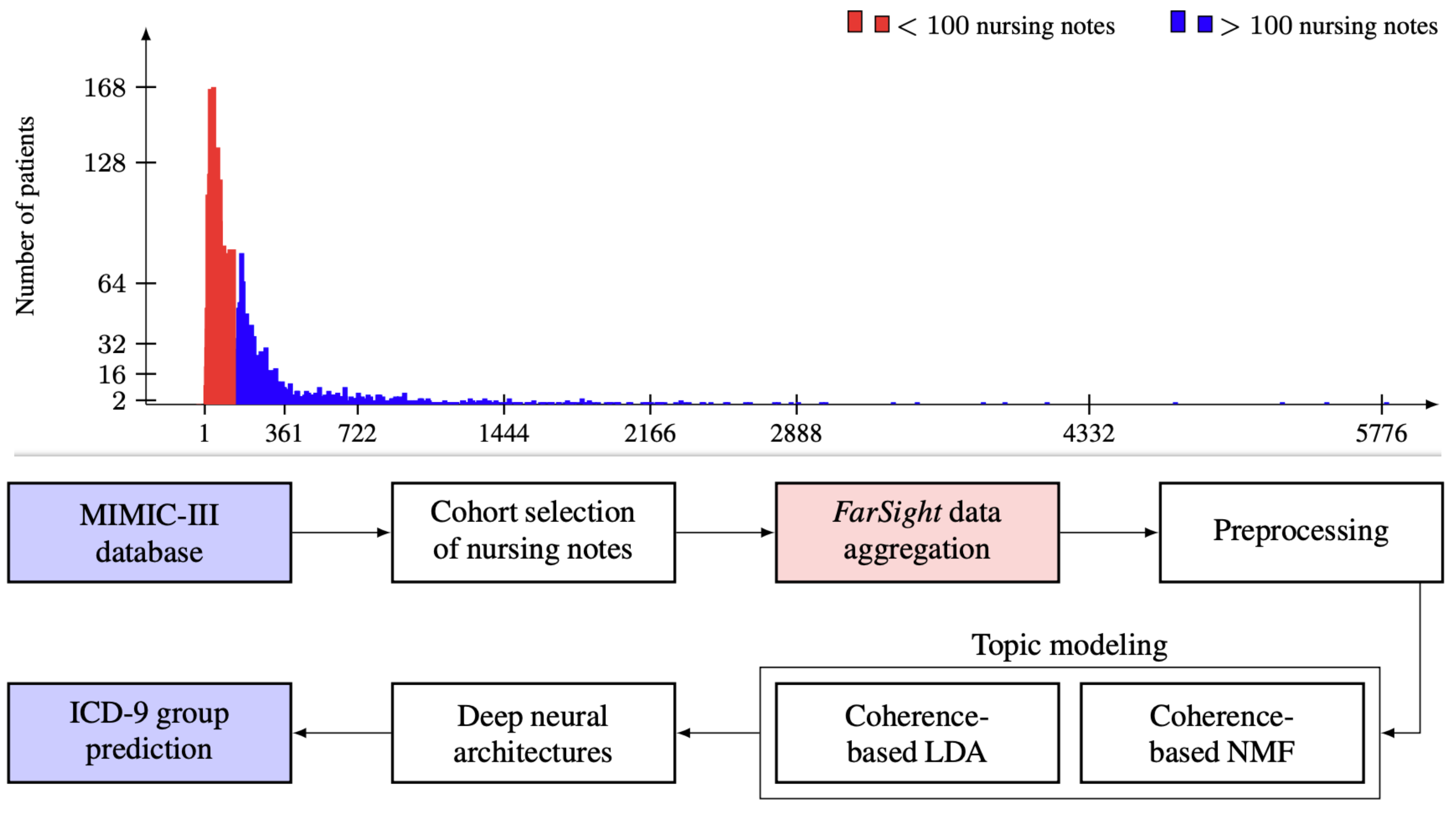
|
Tushaar Gangavarapu, Gokul S Krishnan, and Sowmya Kamath S (2019): Coherence-based Modeling of Clinical Concepts Inferred from Heterogeneous Clinical Notes for ICU Patient Risk Stratification. In Proceedings of the 23rd Conference on Computational Natural Language Learning (CoNLL), ACL (pp. 1012–1022).
abstract
x
Coherence-based Modeling of Clinical Concepts Inferred from Heterogeneous Clinical Notes for ICU Patient Risk Stratification.
In hospitals, critical care patients are often susceptible to various complications that adversely affect their morbidity and mortality. Digitized patient data from Electronic Health Records (EHRs) can be utilized to facilitate risk stratification accurately and provide prioritized care. Existing clinical decision support systems are heavily reliant on the structured nature of the EHRs. However, the valuable patient-specific data contained in unstructured clinical notes are often manually transcribed into EHRs. The prolific use of extensive medical jargon, heterogeneity, sparsity, rawness, inconsistent abbreviations, and complex structure of the clinical notes poses significant challenges, and also results in a loss of information during the manual conversion process. In this work, we employ two coherence-based topic modeling approaches to model the free-text in the unstructured clinical nursing notes and capture its semantic textual features with the emphasis on human interpretability. Furthermore, we present FarSight, a long-term aggregation mechanism intended to detect the onset of disease with the earliest recorded symptoms and infections. We utilize the predictive capabilities of deep neural models for the clinical task of risk stratification through ICD-9 code group prediction. Our experimental validation on MIMIC-III (v1.4) database underlined the efficacy ofFarSight with coherence-based topic modeling, in extracting discriminative clinical features from the unstructured nursing notes. The proposed approach achieved a superior predictive performance when benchmarked against the structured EHR data based state-of-the-art model, with an improvement of 11.50% in AUPRC and 1.16% in AUROC.
url
.pdf
.poster
.bib
x
Coherence-based Modeling of Clinical Concepts Inferred from Heterogeneous Clinical Notes for ICU Patient Risk Stratification.
@inproceedings{gangavarapu-conll-2019,
title = "Coherence-based Modeling of Clinical Concepts Inferred from Heterogeneous Clinical Notes for {ICU} Patient Risk Stratification",
author="Gangavarapu, Tushaar and Krishnan, Gokul S and Kamath S, Sowmya",
booktitle="Proceedings of the 23rd Conference on Computational Natural Language Learning (CoNLL)",
month="Nov",
year="2019",
address="Hong Kong, China",
publisher="Association for Computational Linguistics",
url="https://www.aclweb.org/anthology/K19-1095",
doi="10.18653/v1/K19-1095",
pages="1012-1022"
}
.data
|
conference
healthcare analytics
clinical decision support systems
disease prediction
multi-label classification
natural language processing
deep learning
|
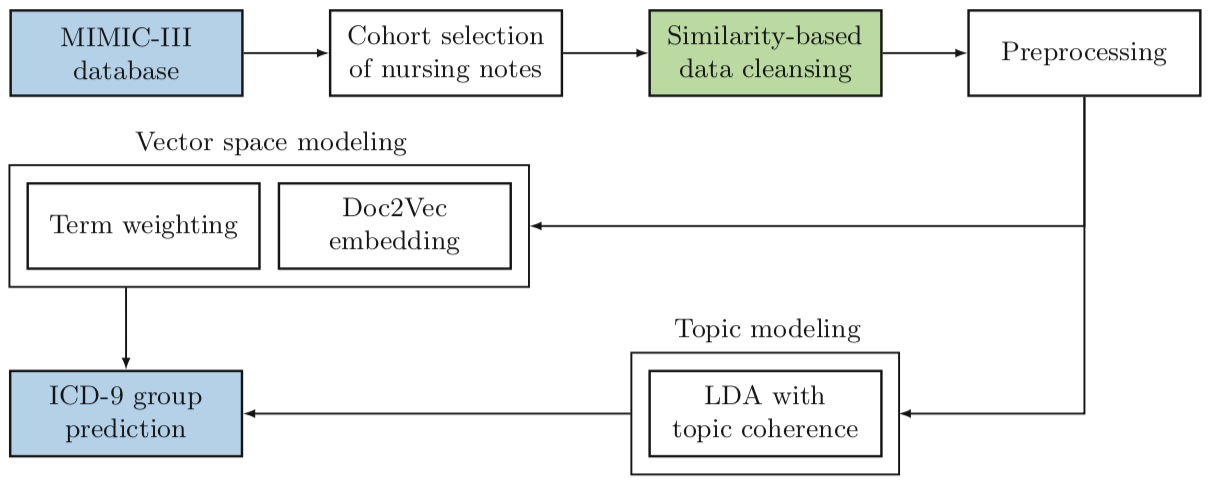
|
Tushaar Gangavarapu*, Aditya Jayasimha*, Gokul S Krishnan, and Sowmya Kamath S (2019): TAGS: Towards Automated Classification of Unstructured Clinical Nursing Notes. In Proceedings of the 24th International Conference on Applications of Natural Language Processing to Information Systems (NLDB), Springer (LNCS 11608, pp. 195–207).
abstract
x
TAGS: Towards Automated Classification of Unstructured Clinical Nursing Notes.
Accurate risk management and disease prediction are vital in intensive care units to channel prompt care to patients in critical conditions and aid medical personnel in effective decision making. Clinical nursing notes document subjective assessments and crucial information of a patient’s state, which is mostly lost when transcribed into Electronic Medical Records (EMRs). The Clinical Decision Support Systems (CDSSs) in the existing body of literature are heavily dependent on the structured nature of EMRs. Moreover, works which aim at benchmarking deep learning models are limited. In this paper, we aim at leveraging the underutilized treasure-trove of patient-specific information present in the unstructured clinical nursing notes towards the development of CDSSs. We present a fuzzy token-based similarity approach to aggregate voluminous clinical documentations of a patient. To structure the free-text in the unstructured notes, vector space and coherence-based topic modeling approaches that capture the syntactic and latent semantic information are presented. Furthermore, we utilize the predictive capabilities of deep neural architectures for disease prediction as ICD-9 code group. Experimental validation revealed that the proposed Term weighting of unstructured notes AGgregated using fuzzy Similarity (TAGS) model outperformed the state-of-the-art model by 5% in AUPRC and 1.55% in AUROC.
url
.pdf (draft)
.slides
.bib
x
TAGS: Towards Automated Classification of Unstructured Clinical Nursing Notes.
@inproceedings{gangavarapu-nldb-2019,
author="Gangavarapu, Tushaar and Jayasimha, Aditya and Krishnan, Gokul S and Kamath S, Sowmya",
title="TAGS: Towards Automated Classification of Unstructured Clinical Nursing Notes",
booktitle="Natural Language Processing and Information Systems",
year="2019",
publisher="Springer International Publishing",
address="Cham",
pages="195-207",
isbn="978-3-030-23281-8"
}
.data
|
conference
healthcare analytics
disease group prediction
natural language processing
risk assessment systems
deep learning
|
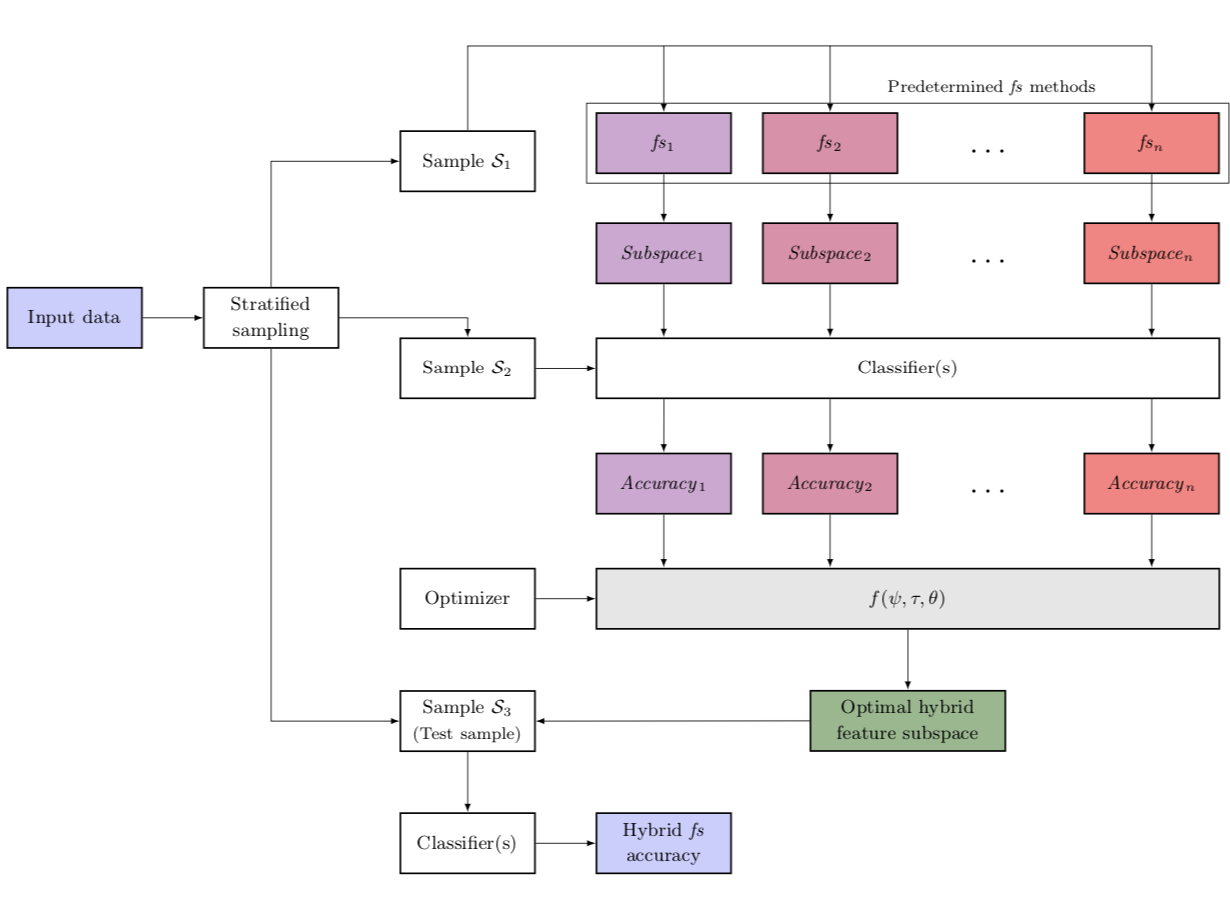
|
Tushaar Gangavarapu and Nagamma Patil (2019): A novel filter—wrapper hybrid greedy ensemble approach optimized using the genetic algorithm to reduce the dimensionality of high-dimensional biomedical datasets. Applied Soft Computing (ASOC), Elsevier 81:105538.
abstract
x
A novel filter—wrapper hybrid greedy ensemble approach optimized using the genetic algorithm to reduce the dimensionality of high-dimensional biomedical datasets.
The predictive accuracy of high-dimensional biomedical datasets is often dwindled by many irrelevant and redundant molecular disease diagnosis features. Dimensionality reduction aims at finding a feature subspace that preserves the predictive accuracy while eliminating noise and curtailing the high computational cost of training. The applicability of a particular feature selection technique is heavily reliant on the ability of that technique to match the problem structure and to capture the inherent patterns in the data. In this paper, we propose a novel filter—wrapper hybrid ensemble feature selection approach based on the weighted occurrence frequency and the penalty scheme, to obtain the most discriminative and instructive feature subspace. The proposed approach engenders an optimal feature subspace by greedily combining the feature subspaces obtained from various predetermined base feature selection techniques. Furthermore, the base feature subspaces are penalized based on specific performance dependent penalty parameters. We leverage effective heuristic search strategies including the greedy parameter-wise optimization and the Genetic Algorithm (GA) to optimize the subspace ensembling process. The effectiveness, robustness, and flexibility of the proposed hybrid greedy ensemble approach in comparison with the base feature selection techniques, and prolific filter and state-of-the-art wrapper methods are justified by empirical analysis on three distinct high-dimensional biomedical datasets. Experimental validation revealed that the proposed greedy approach, when optimized using GA, outperformed the selected base feature selection techniques by 4.17%–15.14% in terms of the prediction accuracy.
url
.pdf (draft)
.highlights
.slides
.bib
x
A novel filter–wrapper hybrid greedy ensemble approach optimized using the genetic algorithm to reduce the dimensionality of high-dimensional biomedical datasets.
@article{gangavarapu-asoc-2019,
author="Gangavarapu, Tushaar and Patil, Nagamma",
title="A novel filter–wrapper hybrid greedy ensemble approach optimized using the genetic algorithm to reduce the dimensionality of high-dimensional biomedical datasets",
journal="Applied Soft Computing",
volume="81",
pages="105538",
year="2019",
issn="1568-4946",
doi="10.1016/j.asoc.2019.105538",
url="http://www.sciencedirect.com/science/article/pii/S156849461930314X"
}
|
journal
biomedical data
genetic algorithm
greedy ensemble
high-dimensional data
hybrid feature selection
parameter optimization
|












